Isolated from a lake in Southern California, Magnetovirga frankelii is a newly identified species of magnetotactic bacterium recently characterized by researchers at BIAM. Beyond expanding our understanding of life, this discovery contributes to biodiversity preservation and opens up exciting possibilities around the bacterium’s ability to navigate using Earth’s magnetic field—a promising trait for developing innovative biotechnologies in medicine and environmental cleanup.
This discovery marks the culmination of a long scientific journey that began in the field, in a location as unique as the bacterium itself. The story starts in 2010, during a sampling mission at the Salton Sea—an hypersaline lake formed accidentally in 1905 following a major flood of the Colorado River. In these extreme saline conditions, researchers succeeded in cultivating an unusual bacterium, temporarily named “SS-5.” After years of dedicated research conducted exclusively at BIAM (within the BEAMM and MEM teams), the strain has now been fully described, characterized, and officially recognized as a new species.
A world first among Gammaproteobacteria
Through genomic, microscopic, and microbial physiology analyses, scientists found that strain SS-5 did not match any known genus and displayed unique features. It is the first cultured magnetotactic bacterium ever identified within the Gammaproteobacteria class.
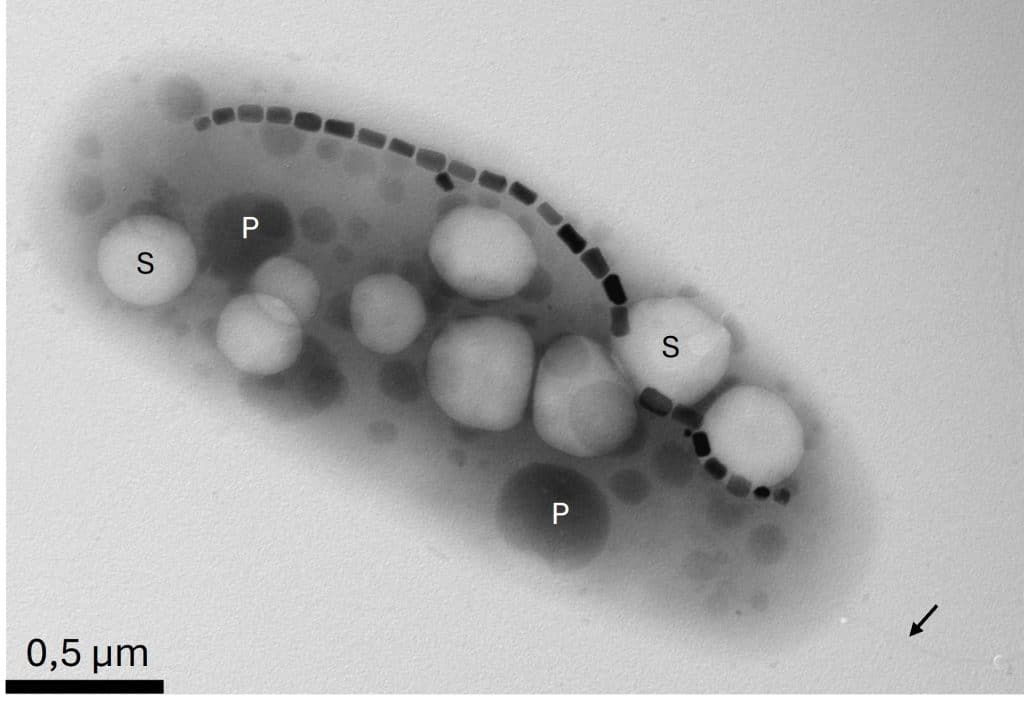
Magnetovirga frankelii produces a chain of magnetosomes—tiny iron oxide crystals—aligned within its cell like an internal compass. Propelled by a single polar flagellum, this microaerophilic bacterium thrives only in the presence of inorganic sulfur compounds.
A new genus, Magnetovirga (from the Latin magneto for magnetism and virga for rod-shaped), was created. The species name, frankelii, honors Richard Frankel, a pioneer in magnetotactic bacteria research. The bacterium is thus officially named Magnetovirga frankelii.
A rigorous process for official recognition
Behind the name lies a stringent scientific process governed by the International Code of Nomenclature of Prokaryotes (ICNP). To be officially named, a bacterium must be i) isolated in pure culture (from a single clonal strain), ii) Thoroughly characterized—morphologically, metabolically, and genetically, iii) deposited in two internationally recognized culture collections—here DSMZ in Germany and JCM in Japan in this case, to ensure lasting access, and iv) described in a peer-reviewed scientific journal. Only after completing these steps, Magnetovirga frankelii becomes an international reference in its field.
A new model for studying organelles in bacteria
Today, Magnetovirga frankelii serves as a model organism at BIAM for studying magnetotaxis and the biogenesis of magnetosomes—fascinating structures that allow bacteria to navigate like a compass.
Two new studies are underway at BIAM to further investigate unique properties of this newly identified species.
Promising impacts for science and society
Beyond the fundamental breakthrough, the discovery and formal naming of this new bacterial species pave the way for potential applications across health, environmental science, and biotechnology—from bioremediation of pollutants to medical imaging, and innovations yet to be imagined.
References
Mila Sirinelli-Kojdinovic, Elsa Turrini, Emma Ropion, Béatrice Alonso, Marine Bergot, Emilie Gachon, Philippe Ortet, Paul Soto-Rodriguez, Caroline Monteil, Christopher Lefevre