press release
Transcription plays a key role within cells by ensuring the copying and amplification of information carried by the genetic material (DNA), in the form of messenger RNAs (mRNAs). Its study is crucial for understanding the functioning of living organisms. Indeed, in both animals and plants, 5 to 6% of genes are involved in regulating this phenomenon, clearly illustrating its biological importance.
Discovered in 1961, mRNA transcription was until now mainly observed on isolated and fixed animal cells, thanks to cutting-edge techniques developed over many years by a team from the Montpellier Human Genome Institute (CNRS/University of Montpellier). The absence of movement in plants has finally made it possible to move on to a new scale of complexity for all the cells of a living organism. Indeed, a team from the Aix-Marseille Institute of Biosciences and Biotechnologies (BIAM CEA/CNRS/AMU)[1] has succeeded in observing and filming in real time the transcription of mRNA in all the cells of a plant.
This work now makes it easier to decipher the mechanisms of many phenomena controlling the development and adaptation of plants in response to the constraints of their environment.
For example, conventional techniques previously showed that plants responded in less than 20 minutes to phosphate inputs (one of their main nutrients). The use of these new approaches has shown that it actually takes only 3 to 5 minutes for the plant to respond to changes in its nutrient environment.
Furthermore, these analyses also revealed significant transcriptional heterogeneity between cells that are supposed to perform identical functions – something scientists had no idea was going on!
How does it work?
(CNRS/University of Perpignan – Via Domitia), Physiological and Pathological Cell Dynamics (CNRS/Institut Pasteur), the Cellular and Plant Physiology Laboratory (CNRS/CEA/INRAE/University of Grenoble Alpes) and the Institute of Human Genetics in Montpellier (CNRS/University of Montpellier).
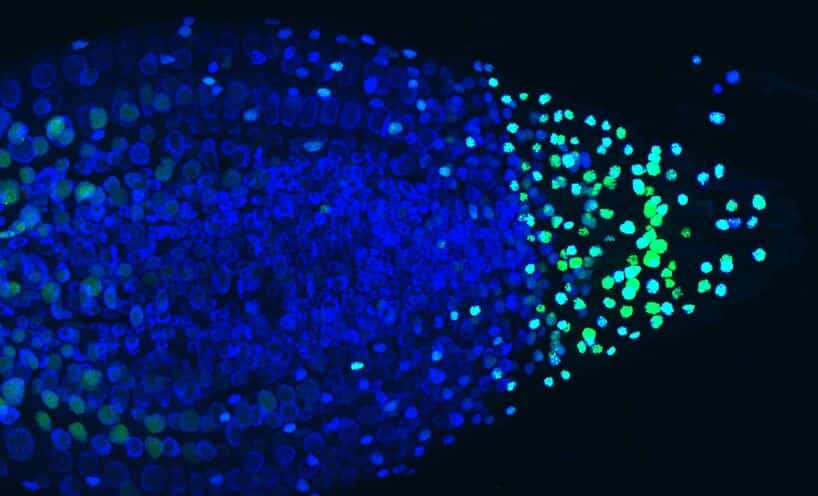
In order to visualise mRNA molecules (i.e. objects from 10 to 500 nm), the scientists used the expression of a viral protein fused to a fluorescent marker. This protein recognises a specific RNA sequence introduced into the plant’s target gene in the gene to be studied. When the latter is transcribed into RNA, it aggregates 256 fluorescent proteins on the mRNA produced. This system makes it possible to directly observe the DNA transcription zones within the cell nucleus (Fig1).
A technical feat
Plants are proving to be an ideal experimental material for studying cells: unlike animal organisms, they are easily immobilised.
Recent technologies such as microfluidics allow the fine control of the nutrition provided to plants, while being compatible with microscopic observations. To do this, a polymer is cast onto a microscope slide. It incorporates fine channels designed to allow root growth and feeding while limiting their movement as much as possible.
The BIAM team combined this tool with high-resolution, high-speed imaging, enabling the acquisition of 200 images in a few seconds. In this way, they were able to access all the cell layers of the root and follow in real time the changes that occur when a plant is supplied with phosphate.
To view the transcript at an unprecedented resolution:
https://imjoy.io/lite?plugin=muellerflorian/hani-ms2:hani-ms2-sample-3
https://imjoy.io/lite?plugin=muellerflorian/hani-ms2:hani-ms2-sample-1
AFFILIATIONS
- 1Aix Marseille Univ, CEA, CNRS, BIAM, UMR7265, SAVE (Signalisation pour l’Adaptation des Végétaux à leur Environnement), Saint-Paul lez Durance, France.
- 2Agroinnovation International-TIMAC AGRO, Groupe Roullier, Saint-Malo, France.
- 3Department of Animal, Plant and Soil Sciences, Australian Research Council Centre of Excellence in Plant Energy Biology, School of Life Sciences, La Trobe University, Bundoora, Victoria, Australia.
- 4UMR5096 CNRS/Université de Perpignan, Laboratoire Génome et Développement des Plantes, Perpignan, France.
- 5Unité Imagerie et Modélisation, Institut Pasteur and CNRS UMR 3691, Paris, France.
- 6MRI, BioCampus Montpellier, CRBM, Univ. Montpellier, CNRS, Montpellier, France.
- 7UMR 5168 CNRS-CEA-INRA-Université Grenoble Alpes, Laboratoire de Physiologie Cellulaire et Végétale, iRIG, CEA-Grenoble, Grenoble, France.
- 8Institut de Génétique Moléculaire de Montpellier, Univ. Montpellier, CNRS, Montpellier, France. edouard.bertrand@igh.cnrs.fr.
- 9Institut de Génétique Humaine, Univ. Montpellier, CNRS, Montpellier, France. edouard.bertrand@igh.cnrs.fr.
- 10Equipe labélisée Ligue Nationale Contre le Cancer, Montpellier, France. edouard.bertrand@igh.cnrs.fr.
- 11Aix Marseille Univ, CEA, CNRS, BIAM, UMR7265, SAVE (Signalisation pour l’Adaptation des Végétaux à leur Environnement), Saint-Paul lez Durance, France. lnussaume@cea.fr.