You are here : Home City of energies > HOME BIAM > Search > IPM > IPM equipment
Our equipments
The Metal Protein Interactions team has a “micro TOF-Q” mass spectrometer (Bruker) with hybrid geometry type Q-TOF (quadripole – Time Of Flight) equipped with an electrospray source (ESI).
The ESI Q-TOF mass spectrometer enables high-precision analyses to be carried out on proteins or protein complexes of high molecular weight as well as on peptides or small molecules. Differents protein characterizations are thus possible :
- Determination of the exact mass of proteins and peptides
- Control of the expression, modification, mutation and labelling of proteins and peptides
- Analysis of gentle proteolysis of proteins
- Study of protein complexes by native mass spectrometry.
Studies of interactions within non-covalent protein/partner complexes, identification of partners and stoichiometry (proteins/proteins – proteins/metals – proteins/cofactors – proteins/DNA) can be carried out by mass spectrometry from samples in liquid phase in a volatile biological buffer. The TOF analyser coupled to an electrospray source allows these measurements of non-covalent complexes owing to a wide range of mass values in m/z (mass on charge). (Leney AC, Heck AJ (2017) Native Mass Spectrometry: What is in the Name? J Am Soc Mass Spectrom. 28:5-13)
Some examples of applications in mass spectrometry
Highlighting the presence of partners by non-covalent mass spectrometry
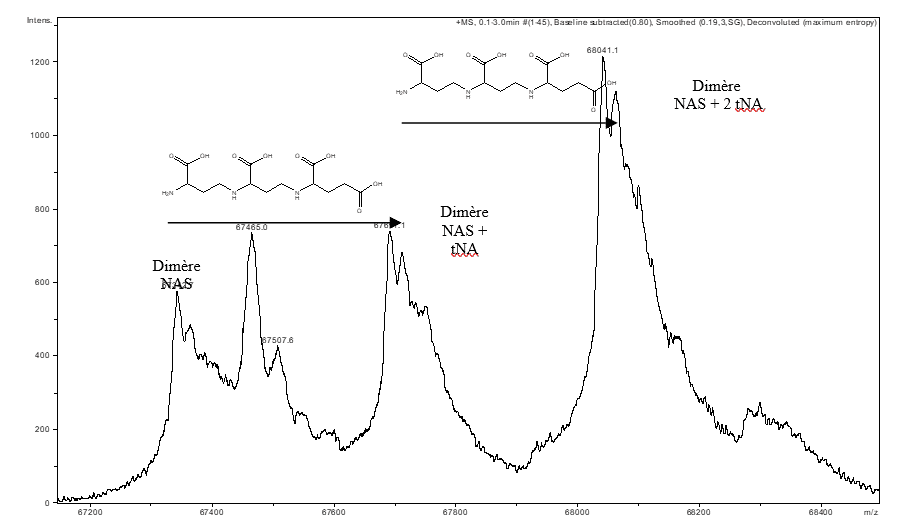
Non-covalent mass spectrometry allows the integrity of complexes maintained by non-covalent interactions to be preserved with the aim of identifying the partners within a complex and their stoichiometry. This type of measurement has made it possible to highlight the presence of a tNA (thermoNicotianamine) ligand within a nicotianamine synthase (NAS) derived from Methanothermobacter thermautotrophicus (Figure A). (Dreyfus & al. PNAS 2009).
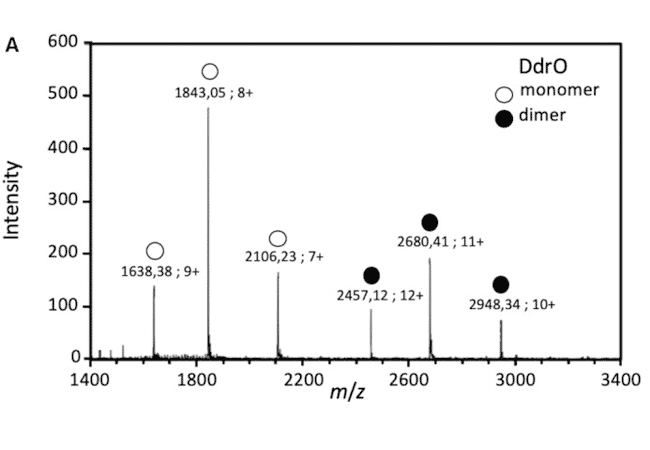
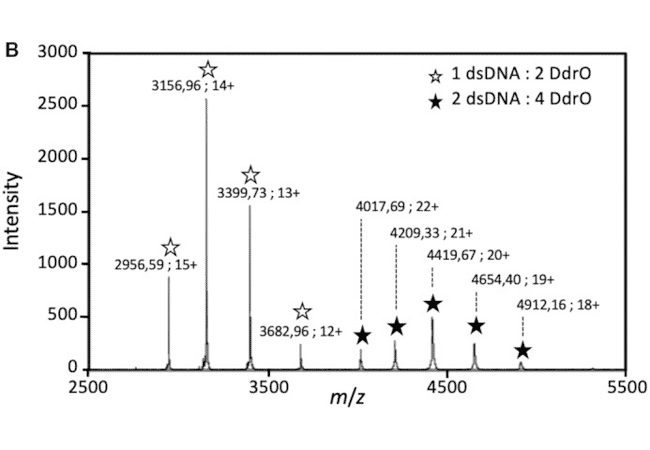
The detection of protein/DNA complexes was obtained by native mass spectrometry as part of a study of DNA repair in deinococcus: interaction between the transcriptional repressor DdrO and DNA (Figure B). (de Groot & al. Nucleic Acids Res. 2019)
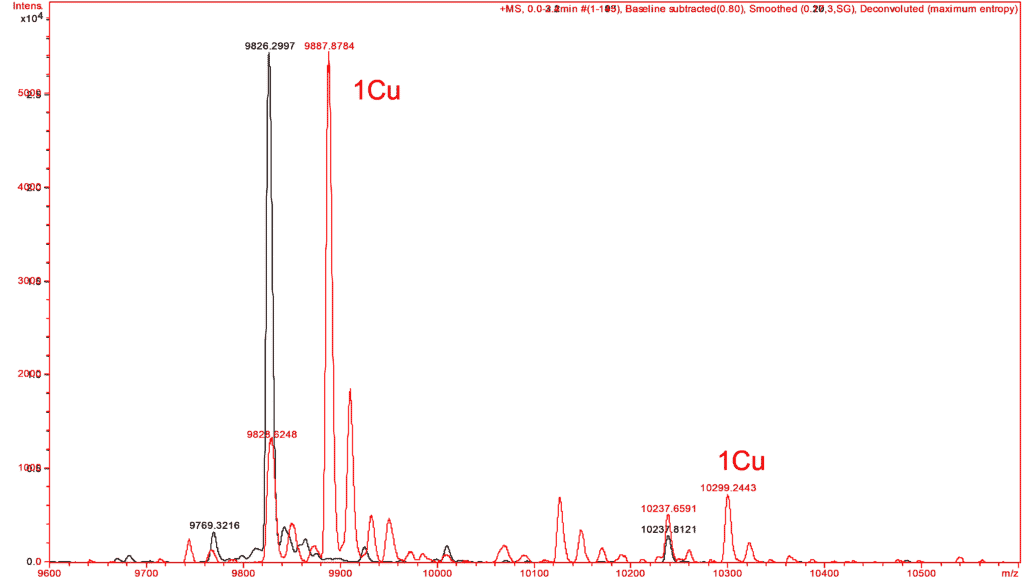
In the same way, similar experiments have made it possible to verify the complexation by copper of a small protein. The mass spectra of this protein are performed in the absence and in the presence of copper reconstituting the expected complex from the apo form of the protein and to verify its metallic partners (Figure C).
Analysis of metal sites in proteins by FTIR spectroscopy in Attenuated Total Reflection (ATR) mode coupled with microdyalisis
The development of a dialysis approach coupled with the acquisition of FTIR spectra in the Attenuated Total Reflection mode allows us to identify the structural changes associated with the binding of a metal in a protein and the nature of the chemical groups involved in the coordination of the metal ion (Gourion-Arsiquaud et al. 2005, Vidaud et al. 2007).
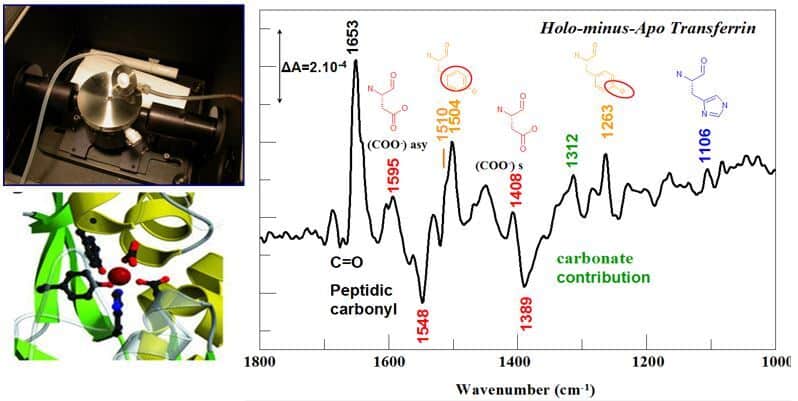
Illustration of the ATR device with connections for the dialysis system, and FTIR difference spectrum corresponding to iron binding ti transferrin with IR bands assigned to the iron ligands. ©Catherine Berthomieu/ BIAM
ATR FTIR spectroscopy can also be used on small proteins or peptides in the form of dehydrated films, to compare apo and metal-bound forms. By determining in particular the presence of monodentate or bidentate carboxylate ligands, and the involvement of phosphoryl groups in the chelation of metals, this approach is very complementary to X-ray absorption spectroscopy approaches (XAS – EXAFS) to characterize metal sites in proteins (Sauge-Merle et al. 2017).
By spectro-electrochemistry in the mind and far IR domains
We have optimized set-ups for coupling electrochemistry and FTIR spectroscopy in the mid and far- IR domains. The mid IR domain provides information on the properties of functional groups of ligands, while the far IR domain allows us to identify the properties of metal-ligand interactions. First results were obtained on cytochrome c and then on Cu,Zn superoxide dismutase (Marboutin et al. 2011). With the model protein azurin, we have shown that it is possible to detect intermolecular hydrogen bonds involving water molecules around 200 cm-1 and the consequences of these bonds on the strength of metal-ligand interactions (Vita et al. 2013). We are currently studying the properties of cytochromes and iron-sulfur centers of ferredoxins (Motomura et al. 2019, Zuccarello et al. 2020).
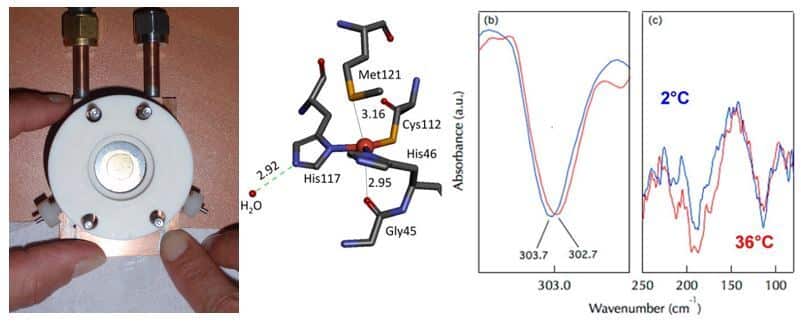
Analysis of photochemical processes in proteins by coupling photochemistry and FTIR spectroscopy, at room or cryogenic temperatures
Light-induced FTIR difference spectroscopy is used to study the photochemical reactions of photosystem II and Fatty Acid Photodecarboxylase (Sorrigue et al. 2020 submitted).
Our equipments consist in a Bruker Vertex 70V and a Bruker 66S FTIR spectrometers equipped with MCT detectors as well as a Si Bolometer, a SensIR ATR, a doubled Nd-Yag laser and a MBT N2-cooled cryostat equipped with IR-transparent windows.
References
- Gourion-Arsiquaud S, Chevance S, Bouyer P, Garnier L, Montillet JL, Bondon A, Berthomieu C (2005) Identification of a Cd2+ and Zn2+ binding site in cytochrome c using FTIR coupled to an ATR micro-dialysis set-up and NMR spectroscopy. Biochemistry 44, 8652-8663.
- Vidaud C, Gourion-Arsiquaud S, Rollin-Genetet F, Albert C, Plantevin S, Pibbe O, Berthomieu C, Quemeneur E (2007) Structural consequences of UO22+ binding to apotransferrin : can this protein account for uranium entry into human cells ? Biochemistry 46, 2215-2226.
- Marboutin L, Petitjean H, Xerri B, Vita N, Dupeyrat F, Flament JP, Berthomieu D, Berthomieu C (2011) Profiling the active site of a cuproenzyme through its far-infrared fingerprint (680-50 cm-1). Angew Chem Int Ed Engl. 50, 8062-8066.
- Vita N, Brubach JB, Hienerwadel R, Bremond N, Berthomieu D, Roy P, Berthomieu C (2013) Electrochemically-induced far-Infrared difference spectroscopy on metalloproteins using advanced synchrotron technology. Anal. Chem. 85, 2891-2898.
- Dalla Bernardina, S, Alabarse F, Kalinko A, Roy, P, Vita N, Hienerwadel R, Berthomieu C, Judeinstein P, Zanotti J-M, Bantignies JL, Haines J, Catafesta J, Creff G, Manceron L, Brubach JB (2014) “New experimental set up for studing nanoconfined water on the AILES beamline at SOLEIL”. Vibrational Spectroscopy, http://dx.doi.org/10.1016/j.vibspec.2014.07.016
- Sauge-Merle S, Brulfert F, Pardoux R, Solari PL, Lemaire D, Safi S, Guilbaud P, Simoni E, Merroun ML, Berthomieu C. (2017) Structural Analysis of Uranyl Complexation by the EF-Hand Motif of Calmodulin: Effect of Phosphorylation. Chemistry. 23, 15505-15517.
- Motomura T, Zuccarello L, Sétif P, Boussac A, Umena Y, Lemaire D, Tripathy JN, Sugiura M, Hienerwadel R, Shen JR, Berthomieu C. (2019) An alternative plant-like cyanobacterial ferredoxin with unprecedented structural and functional properties. Biochim Biophys Acta Bioenerg. 1860(11):148084
- Zuccarello L, Berthomieu C, Boussac A, Brubach JB, Díaz-Moreno I, Díaz Quintana AJ, Hienerwadel R (2020) Protonation of the Cysteine Axial Ligand Investigated in His/Cys c-Type Cytochrome by UV-Vis and Mid- and Far-IR Spectroscopy. J Phys Chem Lett. 11, 4198-4205. doi: 10.1021/acs.jpclett.0c01016.
- Sorigué D, Hadjidemetriou K, Blangy S, Gotthard G, Bonvalet A, Coquelle N, Samire P, Aleksandrov A, Antonucci L, Benachir A, Boutet S, Byrdin M, Cammarata M, Carbajo S, Cuiné S, Doak R B, Foucar L, Gorel A, Grünbein M, Hienerwadel R, Hilpert M, Kloos M, Lane TJ, Légeret B, Legrand P, Li-Beisson Y, Moulin S, Nurizzo D, Peltier G, Schirò G, Shoeman RL, Sliwa M, Solinas X, Zhuang B, Barends TRM, Colletier J-P, Joffre M, Royant A, Berthomieu C*, Weik M*, Domratcheva T*, Brettel K, Vos MH*, Schlichting I, Arnoux P*, Müller P*, Beisson F* (2021) Mechanism and dynamics of photodecarboxylase. Science 372, eabd5687, DOI: 10.1126/science.abd5687
Team manager
Catherine Berthomieu
Contact the team
Key words
Metalloproteins structure-fonction ; Nuclear toxicology ; Biosensors ; Bioremediation ; Metal stress ; Radionuclides