You are here : Home City of energies > HOME BIAM > Search > LGBP > LGBP Research Themes
Research Topics
Our objectives
Stefano CAFFARRI, Rainer HIENERWADEL, Aurélie CREPIN.
A particularity of photosynthetic organisms is their ability to use solar energy to produce complex organic compounds from simple inorganic molecules. In particular, Photosystem II and Photosystem I are important enzymes where the absorption of light and the first steps of its conversion into chemical energy take place. Due to the sessile nature of plants, their photosynthetic apparatus must adapt efficiently to rapid environmental changes such as fluctuations in the quality of incident light. We are developing a multidisciplinary approach in the fields of biochemistry, molecular biology and static and kinetic spectroscopic methods to better understand the particular structures and function of these photosystems. We also aim to clarify the structural adaptations to different light conditions and the defence mechanisms under stressful conditions such as very bright light. For this purpose, we use the different models of photosynthetic organisms present in the laboratory (Arabidopsis, Chlamydomonas,
Physcomitrella, Posidonia et bactéries photosynthétiques).
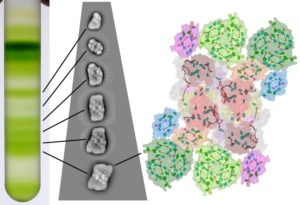
Figure 1: Purification of Photosystem II by ultracentrifugation of membranes solubilised on a sucrose gradient. Photosystem II supercomplexes with a different size of the antennal system migrate to different positions and were visualised by electron microscopy techniques – Credit: Stefano CAFFARRI/ AMU
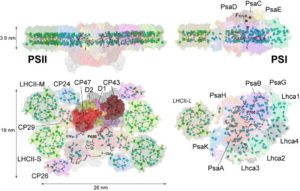
Figure 2: Models of the supercomplex structures of Photosystem II and Photosystem I seen from the lateral and lumenal sides. Key subunits of the PSII core (D1, D2, CP47 and CP43) and its antennal system complexes (LHCII, CP24, CP26 and CP29), and subunits of the PSI (PsaA-B-C-D-E-G-H-K) and its antennal system (Lhca1-4 and LHCII) are shown. The position of most chlorophylls and carotenoids is indicated (for details, see Caffarri et al. Curr. Protein Pept. Sci., 2014) – Credit: Stefano CAFFARRI/ AMU
Our research areas
Structure and organisation of plant photosystems: Purification and structural analysis of the reaction centre, complex cores and supercomplexes of photosystems I and II.
Electron and proton transfer in photosystem II: light- and electrochemical-induced differential FTIR spectroscopy and UV-VIS time-resolved spectroscopy techniques (nanosecond).
Photoprotection of photosystems: regulation of state transitions; role of the PsbS protein; photoprotective role of carotenoids in photosystems and in isolated antennal systems.
Publications Pubmed : here
Chloroplast regulation- the role and molecular mechanisms of ppGpp signalling in plants and algae.
In bacteria the hyperphosphorylated nucleotides guanosine pentaphosphate and tetraphosphate [together referred to as ppGpp] orchestrate a signalling cascade that regulates growth under optimal conditions and in response to environmental stress. ppGpp is also found in the chloroplasts of plants and algae where it has also been shown to accumulate in response to abiotic stress. Studies from our group have shown that ppGpp is an evolutionarily conserved inhibitor of chloroplast gene expression that can regulate photosynthesis, nutrient remobilisation, growth, and immunity in plants and in algae (for review see Field 2018). However, many questions remain on how ppGpp signalling works and how it is integrated into known growth and stress signalling pathways.
Our current and future research is focussed on elucidating the physiological role (what is it for?) and molecular mechanisms (how does it work?) of ppGpp signalling. We work predominantly on the model plant Arabidopsis thaliana where we use molecular genetics, plant physiology, and biochemistry approaches to answer these questions. We are also actively developing new cell biology and synthetic biology techniques to accelerate our research efforts. Our work will reveal the roles that ppGpp signalling appears to play at the nexus of nutrient and energy flows in photosynthetic organisms.
Researchers involved:
Ben Field, Patrice Crété, Benoit Menand, Stefano Caffarri, Christophe Robaglia, Stefano D’Alessandro (visiting scientist), Shanna Romand (PhD), Marwa Mehrez (PhD, visiting), Florent Velay (PhD)
Financement : CHLORO_SAP ANR, SIGNAUX_BIONRJ, SIGNAUX_BIONRJ ANR, G4PLAST ANR
Sur Twitter : @Ben_Field_1, @StefanoDAless11, @RomandShanna
Romand S, Abdelkefi H, Lecampion C, Belaroussi M, Dussenne M, Ksas B, Citerne S, Caius J, D’Alessandro S, Fakhfakh H, Caffarri S, Havaux M, Field B* (2022) A guanosine tetraphosphate (ppGpp) mediated brake on photosynthesis is required for acclimation to nitrogen limitation in Arabidopsis. Elife 10.7554/eLife.75041 Highlight
Harchouni S, England S, Vieu J, Romand S, Aouane A, Citerne S, Legeret B, Alric J, Li-Beisson Y, Menand B* and Field B* (2022), Guanosine tetraphosphate (ppGpp) accumulation inhibits chloroplast gene expression and promotes super grana formation in the moss Physcomitrium (Physcomitrella) patens. New Phytol. https://doi.org/10.1111/nph.18320
Avilan, L., Lebrun, R., Puppo, C., Citerne, S., Cuiné, S., Li‐Beisson, Y., Menand, B., Field*, B. and Gontero*, B. (2021), ppGpp influences protein protection, growth and photosynthesis in Phaeodactylum tricornutum. New Phytol, 230: 1517-1532. https://doi.org/10.1111/nph.17286
Nuclear regulation – the role of Topoisomerase VI in Chromatin and transcription-based stress response.
In plants, most environmental constraints culminate in an oxidative stress originating from the chloroplast. A genetic screen for mutants affected in oxidative stress response led us to identify topoisomerase VI as a key element of the response; this finding highlighted the importance of chromatin-level control in the response of plants to oxidative stress (Šimková et al, 2012). Topoisomerases can regulate the expression of genes by allowing conformational changes of DNA and chromatin, making previously condensed chromatin accessible to the basal transcriptional machinery and transcription factors. Genome-wide gene expression analyses revealed that Topo VI is necessary not only for the full activation of oxidative stress-responsive transcripts, but also for the transcriptional silencing of transposable elements and for sustaining pericentromeric protein coding gene expression. Genome-wide chromatin analyses further revealed that the Topoisomerase VI participates in an insulator-like that prevents heterochromatin spreading into euchromatin islands (Méteignier et al, bioRxiv). We have also identified new interactors of the Topo VI complex that provide hints of possible mechanisms by which Topo VI may act as a regulator of gene expression and function in an insulator-like function required for the organization of the genome into well-delineated transcriptionally active and inactive chromatin domains.
Researchers involved:
Christophe Laloi, Cécile Lecampion, Florent Velay (PhD)
Financement : SLOSAM ANR, ADASOX, CHROS ANR.
Selected publications :
Méteignier LV, Lecampion C, Velay F, Vriet C, Dimnet L, Térèse M, Rougée M, Breuer C, Soubigou-Taconnat L, Sugimoto K, Barneche F and Laloi C (2022) Topoisomerase VI participates in an insulator-like function that prevents H3K9me2 spreading into euchromatin islands. PNAS 119 (27) e2001290119 https://doi.org/10.1073/pnas.2001290119
Bourbousse C, Barneche F and Laloi C (2020) Plant Chromatin Catches the Sun. Front. Plant. Sci. 10(1728). doi: 10.3389/fpls.2019.01728.
Simkova, K., Moreau, F., Pawlak, P., Vriet, C., Baruah, A., Alexandre, C., Hennig, L., Apel, K. And Laloi, C. (2012) Integration of stress-related and reactive oxygen species-mediated signals by Topoisomerase VI in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109(40), 16360-16365. doi:10.1111/j.1365-313X.2011.04825.x.
LGBP : Benoît Menand, Marie-Hélène Montané, Christophe Robaglia, Cécile Lecampion, Manal Boulaiz, Ben Field
Collaborations : Christian Meyer (INRAe, Versailles), Jean-Luc Gallois (INRAe, Avignon), Brigitte Gontero (CNRS, IMM, Marseille), Thierry Desnos (BIAM/SAVE), Yonghua Li-Beisson (BIAM/EBM), Fabien Nogué (INRAe Versailles)
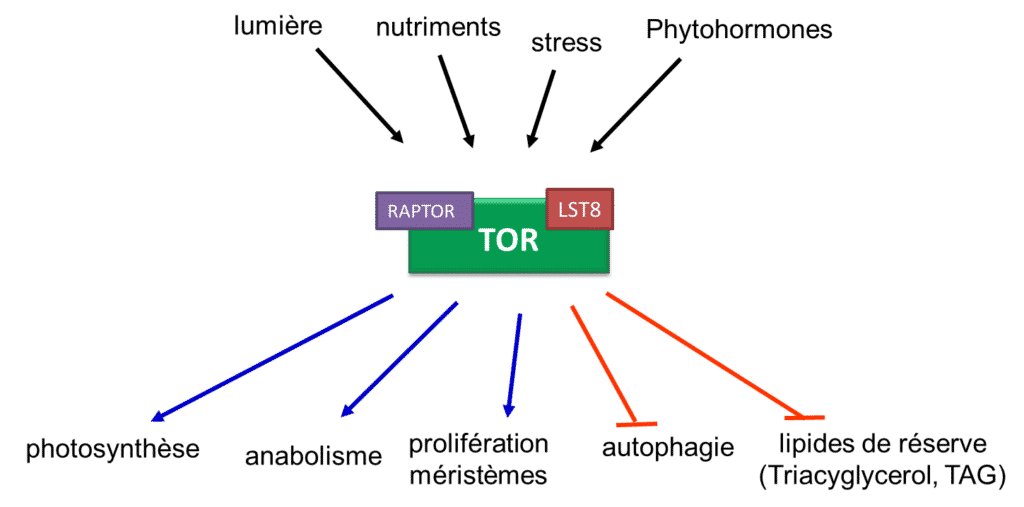
Figure 1: Main functions connected to the TOR signalling pathway in plants (B. Menand/CNRS)
Objectives:
The adaptation of plants to their environment requires fine regulation of metabolism and growth processes by highly connected signalling pathways. The signalling pathway involving the TOR (Target Of Rapamycin) kinase is conserved during the evolution of eukaryotes and plays a central role in regulating the response to the whole range of environmental perceptions, from various nutrient deficiencies to the response to pathogens. It is therefore a pure model for studying the integration of multistress in plants, a major issue for agriculture (Fig 1.).
Our aim is to identify new components of the TOR signalling pathway in plants (in particular the model plant Arabidopsis thaliana) in order to understand how this ancestral signalling pathway has integrated plant specificities, such as photosynthesis, during evolution. We are also working on algae to understand how modulation of this signalling pathway can improve productivity of triacylglycerols (TAGs), reserve lipids that are precursors to biofuels.
Key highlights
In order to identify new elements of the TOR signalling pathway, we performed a pharmacogenetic screen. This involves selecting Arabidopsis mutants that are either hypersensitive or resistant to TOR inhibition. For this purpose, we use competitive ATP inhibitors of TOR which we have shown to be selective in plants (Fig. 2 and 3).
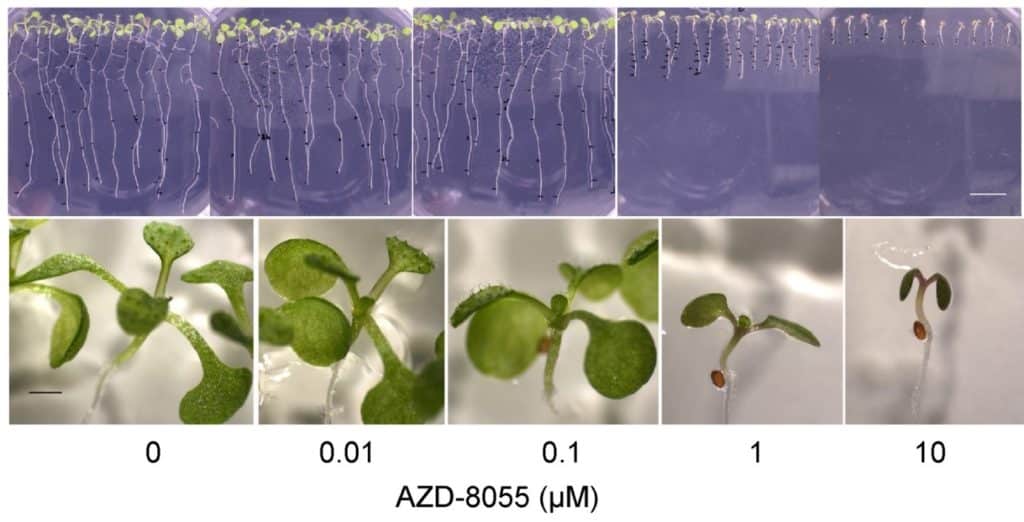
Figure 2: Dose-dependent inhibition of plant growth by AZD-8055, a selective ATP-competitive TOR inhibitor. Three-day-old Arabidopsis thaliana plants were transferred to medium containing the inhibitor and grown for 6 days. Photos of plants growing vertically on the surface of the medium (upper part) and of the aerial part (lower part) on AZD-8055. Credit: B. Menand/CNRS and M.-H. Montané/CEA, J ; Exp. Bot. 2013
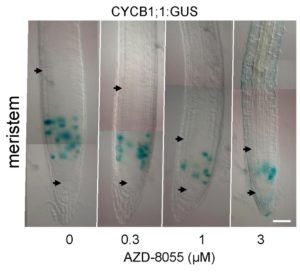
Figure 3: A TOR active site inhibitor (AZD-8055) reduces the size of the root meristem and the proliferation zone but not the duration of the G2 phase of the cell cycle. Photos of root tips under DIC microscopy with the meristematic zone delimited by the arrows. CYCB1;1::GUS expression (blue) marks cells in G2 phase of the cell cycle. Credit: B. Menand/CNRS and M.-H. Montané/CEA
This strategy allowed us to identify a downstream target of TOR, the DYRK kinase YAK1, as an essential regulator of meristem activity. (Fig. 4). We found that yak1 loss-of-function mutants are resistant to TOR inhibition because YAK1 is an essential intermediate of TOR-dependent regulation of the cell cycle that thereby blocks cell proliferation.
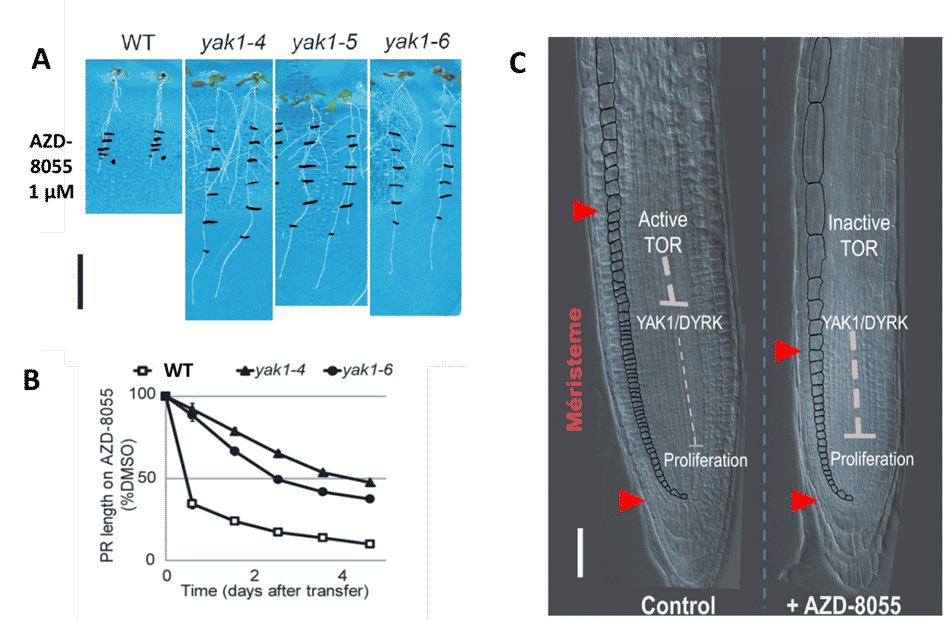
Figure 4: Schematic representation of the role of the TOR-YAK1 axis in the regulation of meristematic activity. B. Menand/CNRS, after Barrada et al, Development 2019. YAK1 is a repressor of cell proliferation in the meristem. When TOR is active, it inhibits YAK, which promotes proliferation and growth, whereas when TOR is inhibited, YAK1 becomes active and blocks cell divisions.
On the other hand, we showed that TAG productivity by algae, treated with a dose of TOR inhibitor that does not block cell proliferation, was 3 to 4 times higher than in nitrogen-deficient algae (Fig.5; Prioretti et al 2017). This increase in lipid productivity is associated with a moderate slowing of proliferation and controlled by partial inhibition of TOR. These conditions constitute a ‘get-fat growth’ regime allowing growth and cell division by accumulating TAGs. This demonstrates that modulation of the TOR pathway can increase cell productivity of TAGs.
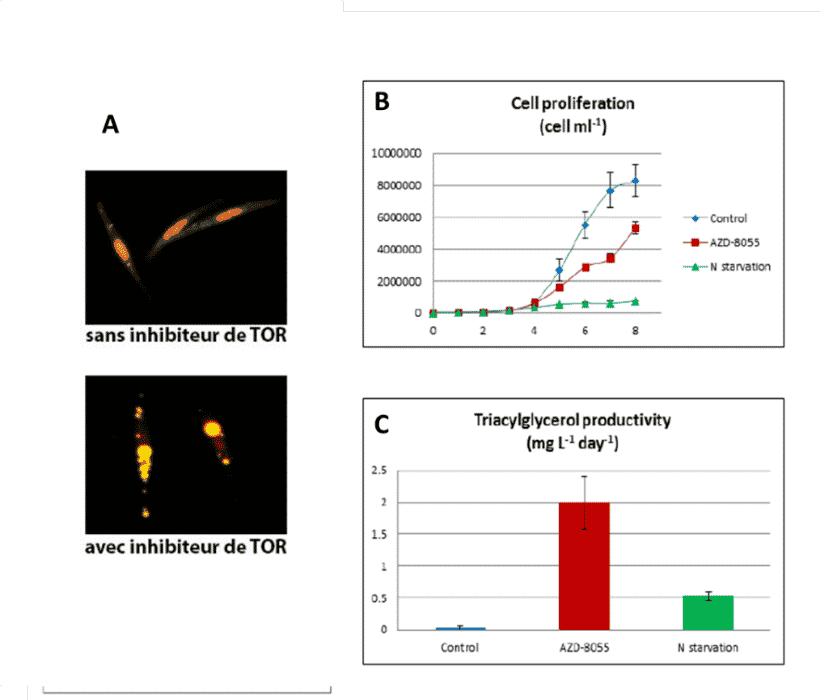
Figure 5: Accumulation of lipid droplets and productivity of reserve lipids, Triacyglycerols, (TAGs) by P. tricornutum diatoms treated with 2 µM of the TOR inhibitor AZD-8055, a concentration that reduces proliferation by 50%. A, Lipid droplets revealed with a fluorescent marker. B, Effect of the TOR inhibitor at a controlled concentration giving a rate of proliferation of up to 50%.
Perspectives
We will continue the analysis of new mutants isolated by the pharmacogenetic screen in order to better understand the functioning of the TOR pathway in plants. In our future analyses, we will distinguish the functions of TOR associated with cell proliferation from those associated with physiology and particularly photosynthesis, for example by looking at nucleus-chloroplast signalling mechanisms (link to research theme 1). We have recently secured funding for a new ANR project (TOR-DYRKcontol link to the project) which aims to decipher the interactions between TOR and DYRK kinases in the control of growth and lipid synthesis in plants and algae. The use of both plants and algae will allow us to transfer knowledge from one species to another, which should also shed light on the evolutionary aspects of TOR signalling. Furthermore, we are interested in the role of the TOR pathway in the establishment of plant resistance to certain viruses in the context of agronomic applications (Ouibrahim et al 2015).
Team manager
Key words
Bioenergy, photosynthesis, photosystems, response to light, response to stress, signalling, Arabidopsis, Chlamydomonas, moss, electron transfer, chloroplast regulation, photosymbiosis, ppGpp, Topoisomerase, chromatin, sea grass, storage lipids, Target of Rapamycin (TOR)